Abstract
Nodular lymphocyte predominant Hodgkin lymphoma (NLPHL) comprises 5% of all Hodgkin lymphomas (HL). Its biology remains poorly characterized. Like classical HL (cHL), it contains minimal malignant cells embedded within a T cell rich intra-tumoral microenvironment (TME). Unlike cHL, it can transform to diffuse large B cell lymphoma (DLBCL). Immune-checkpoint blockade is effective in cHL but has minimal activity in DLBCL. No data is currently available regarding the potential to reactivate host anti-tumoral activity via immune-checkpoint blockade in NLPHL.
Diagnostic FFPE samples from 49 NLPHL patients retrospectively collected from 4 Australian centres were interrogated. Inclusion criteria were sample availability and centrally confirmed histological NLPHL. Characteristics were in line with the literature: median age 45 years, range 13-82 years; F:M 1:3.5; stage I/II 55%, III/IV 35% (10% stage unknown) with the majority of cases were of immuno-architectural types A or C. RNA was digitally quantified using the NanoString 770-gene PanCancer Immune panel. Multi-spectral immunofluorescent (mIF) microscopy, plasma soluble PD-1 quantification, cell sorting, T cell receptor (TCR) repertoire analysis and functional immuno-assays were also performed. Results were compared with samples from 38 cHL and 35 DLBCL patients.
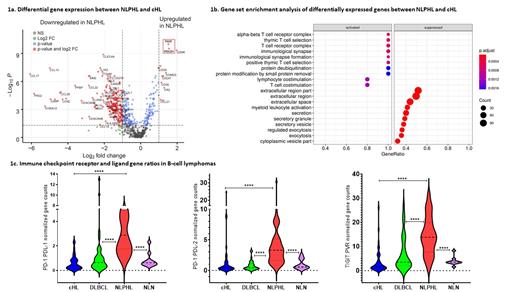
We initially compared gene expression of NLPHL and cHL, looking for molecular similarities and differences. Ten non-lymphomatous nodes (NLN) were included as controls. Unsupervised clustering showed all but 3 NLPHL cases segregated from the cHL cluster. All NLN congregated in a discrete sub-cluster. As expected, RNA analysis showed significant enrichment for CD20 in NLPHL and CD30 in HL. Volcano plots (Fig. 1a), corrected for false-discovery showed marked variation in gene expression. For NLPHL (vs. cHL) there were 105 upregulated and 337 down regulated genes. Strikingly, the most significantly differentially over-expressed genes in NLPHL were all T cell related (CD247: CD3 zeta chain; CD3D: CD3 delta chain; GZMK: granzyme K; EOMES: marker of CD8 + T cell tolerance; and the immune checkpoints PDCD1: encodes for PD-1; and TIGIT). CD8B expression was increased in NLPHL. For cHL, the most over-expressed genes included macrophage-derived chemokines CCL17 and CCL22. Gene set enrichment analysis revealed activation of the PD-L1 expression and PD-1 checkpoint pathway and 9 of the top 10 Gene Ontology (GO) term enrichment scores involved lymphocyte signalling in NLPHL (Fig. 1b). To better appreciate the impact of the relevant immune checkpoints on their signalling axis, we compared gene ratios for PD-1 and TIGIT receptors with their ligands (PD-L1/L2 and PVR, respectively). NLPHL showed the highest enrichment ratios of these signalling pathways vs. cHL, DLBCL and NLN (Fig. 1c).
Although it is known that CD4 +PD-1 +T cells form rosettes around NLPHL cells, the differential cellular localization of immune proteins has not been compared between HL entities. Using mIF, the proportion of intra-tumoral PD-1 + was markedly higher for CD4 + (~7-fold; p<0.0001) and CD8 + (~5-fold; p<0.001) T cells in NLPHL. However, the proportion of T cells expressing LAG3 was similar. Soluble PD-1 was elevated for both NLPHL and cHL, indicating circulating blood is influenced by the TME. For both HL entities over 80% of circulating CD4 + and CD8 + T cells expressed PD-1 alone or in combination with TIGIT. TCR repertoire analysis of sorted T cell subsets showed large intra-tumoral clonal T cell expansions were also detectable in circulating T cells. T cell clones were predominantly PD1 +CD4 + T cells in both HL types.
Finally, we developed a functional assay using PD-L1/PD-L2 expressing NLPHL and cHL cell lines. These were co-cultured with genetically engineered PD-1 +CD4 + T cells that express a luciferase reporter. Similar levels of heightened T cell activation were seen with immune-checkpoint blockade for both HL entities, indicating that immune-checkpoint inhibition may also be of benefit in NLPHL.
In conclusion, our multi-faceted analysis of the immunobiological features of the TME in NLPHL, provides a compelling rationale for early phase clinical studies that incorporate immune-checkpoint blockade in NLPHL.
Hawkes: Bristol Myers Squib/Celgene: Membership on an entity's Board of Directors or advisory committees, Research Funding; Specialised Therapeutics: Consultancy; Merck KgA: Research Funding; Merck Sharpe Dohme: Membership on an entity's Board of Directors or advisory committees; Novartis: Membership on an entity's Board of Directors or advisory committees; Antigene: Membership on an entity's Board of Directors or advisory committees; Regeneron: Speakers Bureau; Janssen: Speakers Bureau; Gilead: Membership on an entity's Board of Directors or advisory committees; Astra Zeneca: Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau; Roche: Membership on an entity's Board of Directors or advisory committees, Other: Travel and accommodation expenses, Research Funding, Speakers Bureau. Swain: Janssen: Other: Travel expenses paid; Novartis: Other: Travel expenses paid. Keane: BMS: Research Funding; Gilead: Membership on an entity's Board of Directors or advisory committees; Janssen: Consultancy; Karyopharm: Consultancy; MSD: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Roche: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees. Talaulikar: Takeda: Honoraria, Research Funding; Amgen: Honoraria, Research Funding; Jansenn: Honoraria, Research Funding; Roche: Honoraria, Research Funding; EUSA Pharma: Honoraria, Research Funding. Gandhi: janssen: Research Funding; novartis: Honoraria.